AnABlast Web Help
AnABlast
is an algorithm to discover signals of protein-coding sequences
within genomic regions. You can analyze a short nucleotide
sequence (up to 25Kb in length or up to 1Mb if you upload the
Blast report). It highlights genomic regions with stacked
non-significant alignments (protomotifs) which would represent
present or ancient protein-coding sequences. It allows to
discover new genes in bacteria or exons in eukaryotic organisms.
Input form
You only have to paste your nucleotide sequence or upload it as a FASTA file, and click on submit. The execution time depends on the length of the input sequence: approximately 1 minute per Kb.
To achieve AnABlast results in a shorter time you can optionally upload a BLAST output file. To do that, you will need to run previously Blast+ program in this way:
$ wget ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/uniref/uniref50/uniref50.fasta.gz
$ gzip -d uniref50.fasta.gz
$ makeblastdb -in uniref50.fasta -dbtype prot
$ blastx -db uniref50.fasta -query sequence.fasta -evalue 50000 -outfmt '6 sseqid qseqid qstart qend evalue bitscore qframe' -matrix BLOSUM90 -seg no -max_target_seqs 10000000
The sensitivity score allow to perform more sensitive predictions (though less specific) changing to bit-score=28, or more specific predictions (though less sensitive) changing to bit-score=35.
Results
The results are shown on a
genome browser (JBrowse)
and are divided in two sections:
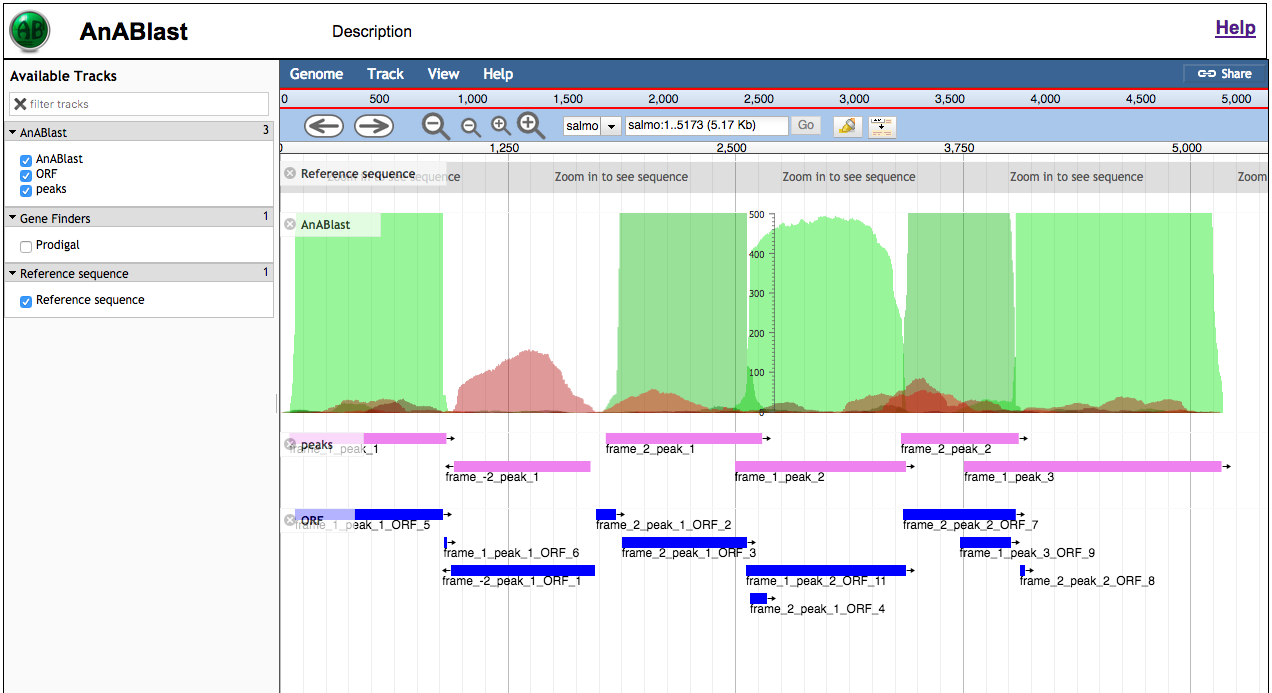
The left block has track
names that you can activate/inactivate:
Reference sequence: Track with the nucleotide sequence.
AnABlast: piling up
of protomotifs throughout the query sequence. In green color
those from the forward strand, and in red color those from the
reverse strand. Commonly, peaks of pile up protomotifs
match with protein-coding regions.
peaks: significant protomotif accumulation (track highligthed in pink). If you click on a peak you will have:
-
Primary Data: name, type, score (peak height), position and
length.
-
Attibutes: Amino acids sequence, frame, peak ID, sequence id,
source, number of stop codons inside the peak, nucleotide
sequence and subfeatures.
-
Subfeatures: functional annotation predicted by Sma3s annotator,
and the best hit blast.
ORF: open reading frames (longer than five aa) found in peak regions (highligthed in blue).
Gene finders: ab initio gene predictions using Prodigal (for prokariotic sequences) and Augustis (for eukaryotic sequences).
The right block shows all the tracks described above and allows you to move along the sequence just dragging the mouse and zoom out or in (so, you can see the nucleotides of the sequence). You can also extract your sequence of interest by zooming to it, clicking on the header "Reference sequence" and selecting "save track data".
References
Rubio A, Casimiro-Soriguer CS, Mier P, Andrade-Navarro MA, Garzón A, Jimenez J, Pérez-Pulido AJ (2019) AnABlast: Re-searching for Protein-Coding Sequences in Genomic Regions. Methods in molecular biology (Clifton, N.J.). 2019; 1962:207-214.
Jimenez J, Duncan CD, Gallardo M, Mata J, Perez-Pulido AJ (2015) AnABlast: a new in silico strategy for the genome-wide search of novel genes and fossil regions. DNA Res. 2015 Oct 21.