The nucleotide sequence against which you wish to find miRNAs produced from a genome. It can be an mRNA sequence, a CDS from a gene, or a sequence coding for several proteins (e.g., a virus genome). This sequence can only have nucleotide letters (ACGTU) and blank spaces.
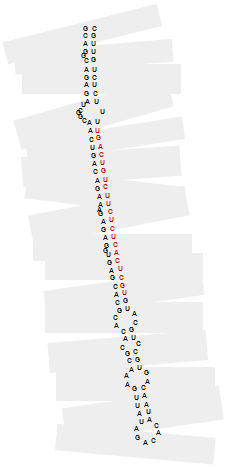
A miRNA is initially transcribed from a genome as an RNA sequence with a stem-loop secondary structure. This is known as the miRNA precursor. It normally has more than 100 nucleotides (see figure).
The miRNA precursor coming from a genome is processed by an enzymatic complex, and only a sequence (from the stem of the structure) of aproximately 20 nucleotides is conserved. This is the mature miRNA, and it undertakes the biological functions of the miRNA (shown as the red sequence in the figure).
This refers to the reference genome where miRNAs against the query sequence are searched for. The complete genomes are downloaded from the NCBI database, and the BACs of organisms without complete genomes come from specific sequencing projects.
This is used by semirna to uniquely identify the predicted miRNA. It is formed by a series of information separated by underscores (_) as follows:
'query name'_'[pm]target position in query sequence'_'[pm]miRNA position in genome'_'genome chromosome number'
where p and m corresponds to plus or minus sequence strand.
This shows the entire sequence of the predicted precursor. The mature miRNA is highlighted in red, in order to show exactly where it is coming from. Additionally, the precursor structure can be shown by clicking on the 'view structure' link.
This is the free energy in kcal/mol of the stem-loop precursor structure. Lower (more negative) values show more stable structures.
The number of the chromosome on which the precursor sequence has been found.
The position of the precursor in the chromosome of subject genome. If it is on the minus strand, the format will be: 'Complement (position1..position2)', with these positions being the positions with respect to the plus strand.
It shows the complementarity alignment of the predicted mature miRNA sequence and the target of the query sequence, both with corresponding positions. The identical positions show alignments between (A-U) and (G-C) in addition to (G-U).
The relative position of the precursor in the subject genome. If the position is intergenic, the surrounding genes are shown, and if it is within a gene then the gene is shown. In addition, a link to the GBrowse is included with the precursor position in a genomic map.
Free energy in kcal/mol of the miRNA-target alignment. Lower (more negative) values show more stable structures.
Quality value for the predicted miRNA precursor. It is the ratio between the free energy value and the nucleotide length of the precursor. Higher values correspond to more stable structures. The default semirna value is 0.35, which was proven with known miRNAs in the miRBase database.