Getting Started
Introduction
Currently, analysis and annotation of genomes focuses primarily on protein-coding genes, using homology searches based on genomic techniques and expression profile definitions based on transcriptomic methods. However, it is becoming ever more obvious that there are many non-coding RNA genes with functions that must also be clarified if we want to understand the complex machinery of living organisms. Historically, identification of these genes and genomic regions has been quite erratic, and only certain classes of RNAs with very strongly conserved sequences and/or structures have been identified. MicroRNAs are noncoding molecules, generally between 20 and 25 nucleotides in length, and are initiators of gene silencing, a crucial process for developmental regulation, virus evolution and defence. Gene silencing works through specific base-pairing with target molecules (usually mRNA). Increasing numbers of miRNAs have been discovered in recent years, with a high diversity of biogenesis and functions. It has been opined that no more miRNAs can be found by current wetlab techniques, leading to the need for computational tools to aid in their discovery.
Here we present semirna, a web server for the detection of miRNAs from a given genome against a query sequence. The algorithm begins with a highly tuned BLAST search in order to identify where the query sequence might show complementarity to the chosen genome. Since gene silencing begins through the small RNA pairing with the mRNA, a region of high homology between the two sequences would indicate an area where the miRNA might pair with the mRNA. Each result of the BLAST report is then processed individually by the rest of the algorithm to see whether it indicates a potential small RNA. It must pass through a series of filters, which take into account particular characteristics of the known miRNA and how it is formed, creating a rule based system. Additionally, the program uses existing programs from the Vienna package to add further filters: RNAfold (folding the putative miRNA precursor), RNAcofold (calculating the free energy of the mature miRNA-target binding) and RNAplot (building a picture of the miRNA folded precursor). All of this results in a low number of false-positive results.
We calculate an additional quality coefficient based on experimentally verified miRNA precursors taken from miRBase which takes into account their length and free energy, and remove low complexity regions from the genomes in order to avoid false positive miRNAs coming from simple nucleotide repeats.
Parameter form
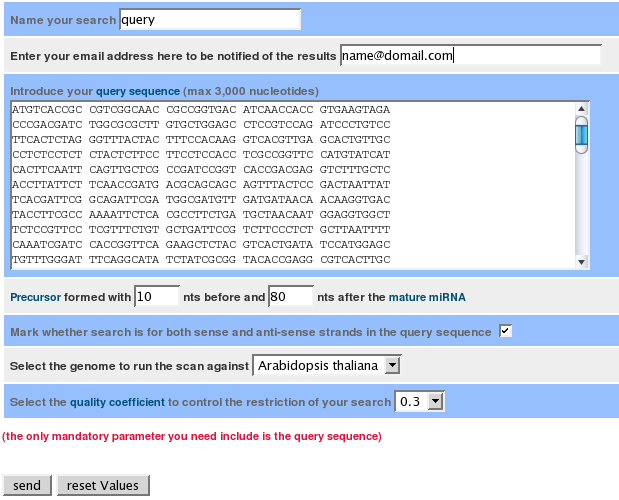
The main page of semirna has a form with the input parameters. From here, you can access help from the top menus or links through the 'Glossary of terms' link. The only mandatory parameter you must include is the query sequence.
Search name. The name for your analysis. If it is not included, the name is fixed at 'query'.
E-mail. If it is included, you will receive an e-mail with the result link when the analysis finishes.
Query sequence. The nucleotide sequence against which you want to find miRNAs. It can be an mRNA sequence, a CDS from a gene, or a sequence coding for several proteins (e.g., a virus genome). This sequence must have only nucleotide letters (ACGTU) and blank spaces.
miRNA flanking nucleotides. Initially, miRNAs exist as precursor molecules, sequences of approximately 100 nucleotides folded into a given secondary structure. You must give a maximum number of nucleotides before and after the mature miRNA to form the precursor. By default, the number is 10 and 80 before and after the mature miRNA, respectively, since this gives the best empirical results, but sometimes you can obtain new results or reinforce previous ones by changing the parameters to a maximum of 100 and 300.
Query sequence strand. If the given query sequence is a coding and sense sequence, you should mark this option in order to avoid searching for miRNAs against the antisense sequence. If you do not know the sense strand from your sequence or it is a multicoding sequence (e.g., a virus sequence), this option should not be marked. The final results will indicate against which strand the miRNA has been found.
Genome. The genome where you want to find miRNAs. The predicted miRNAs would recognise segments from the query sequence.
Quality coefficient. Quality value for the predicted miRNA precursors. It is the quotient of the free energy value and the nucleotide length of the precursor. Higher values correspond to more stable structures. The semirna default value is 0.35, which has been proven with known miRNAs in the miRBase database.
When you click on the 'send' button, a page displaying the current progress will appear. After a variable amount of time, you will have the final results.
Predicted miRNA map
TThe results are initially given as an image map. A scaled ruler represents the length of the query sequence, and the predicted miRNAs are shown below. Each coloured bar is a miRNA, which appears together with its quality coefficient and its identifier. miRNAs with higher coefficient values are darker coloured. To obtain tables with detailed information about the predicted miRNAs, you must click on a specific bar.
Additional result files are also provided by clicking on the links below:
miRNAs in FASTA format. Show all predicted mature miRNA sequences in FASTA format.
Precusors in FASTA format. Show all predicted precursor miRNA sequences in FASTA format.
Predictions in GFF format. Show a GFF format file of the predicted miRNAs, which can be useful for genome viewers.
Multiple alignment of miRNAs. Show a multiple alignment of predicted and known miRNAs all together. The identifiers have links to both miRBase and the detailed result tables from your analysis.

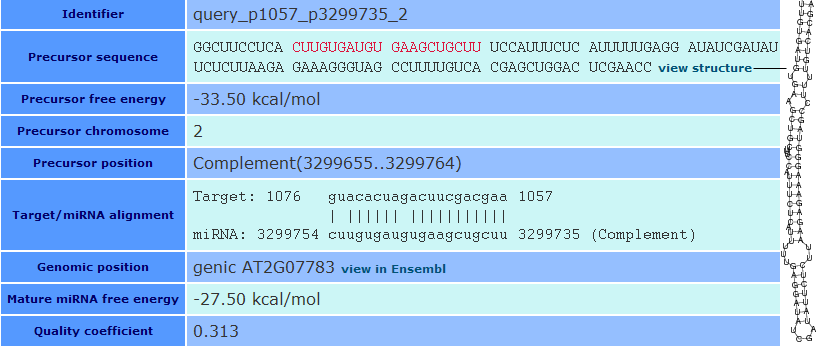
Detailed result tables
When you select a specific predicted miRNA from the image map, semirna shows a detailed table or record with all the parameters from the predicted miRNAs ordered in different lines or fields. To obtain a complete description for each field, please go to the Glossary of Terms.